BioKIT, a versatile toolkit for processing or conducting analyses on alignments, coding sequences, fastq files, and genome files.
If you found BioKIT useful, please cite BioKIT: a versatile toolkit for processing and analyzing diverse types of sequence data. Genetics. doi: 10.1093/genetics/iyac079.
Quick Start
1) Installation
To install using pip, we strongly recommend building a virtual environment to avoid software dependency issues. To do so, execute the following commands:
# create virtual environment
python -m venv .venv
# activate virtual environment
source .venv/bin/activate
# install biokit
pip install jlsteenwyk-biokit
Note, the virtual environment must be activated to use biokit.
After using BioKIT, you may wish to deactivate your virtual environment and can do so using the following command:
# deactivate virtual environment
deactivate
Similarly, to install from source, we strongly recommend using a virtual environment. To do so, use the following commands:
# download
git clone https://github.com/JLSteenwyk/BioKIT.git
cd BioKIT/
# create virtual environment
python -m venv .venv
# activate virtual environment
source .venv/bin/activate
# install
make install
To deactivate your virtual environment, use the following command:
# deactivate virtual environment
deactivate
Note, the virtual environment must be activated to use biokit.
To install via anaconda, execute the following command:
conda install -c jlsteenwyk jlsteenwyk-biokit
Visit here for more information: https://anaconda.org/JLSteenwyk/jlsteenwyk-biokit
2) Usage
Get the help message from BioKIT:
biokit -h
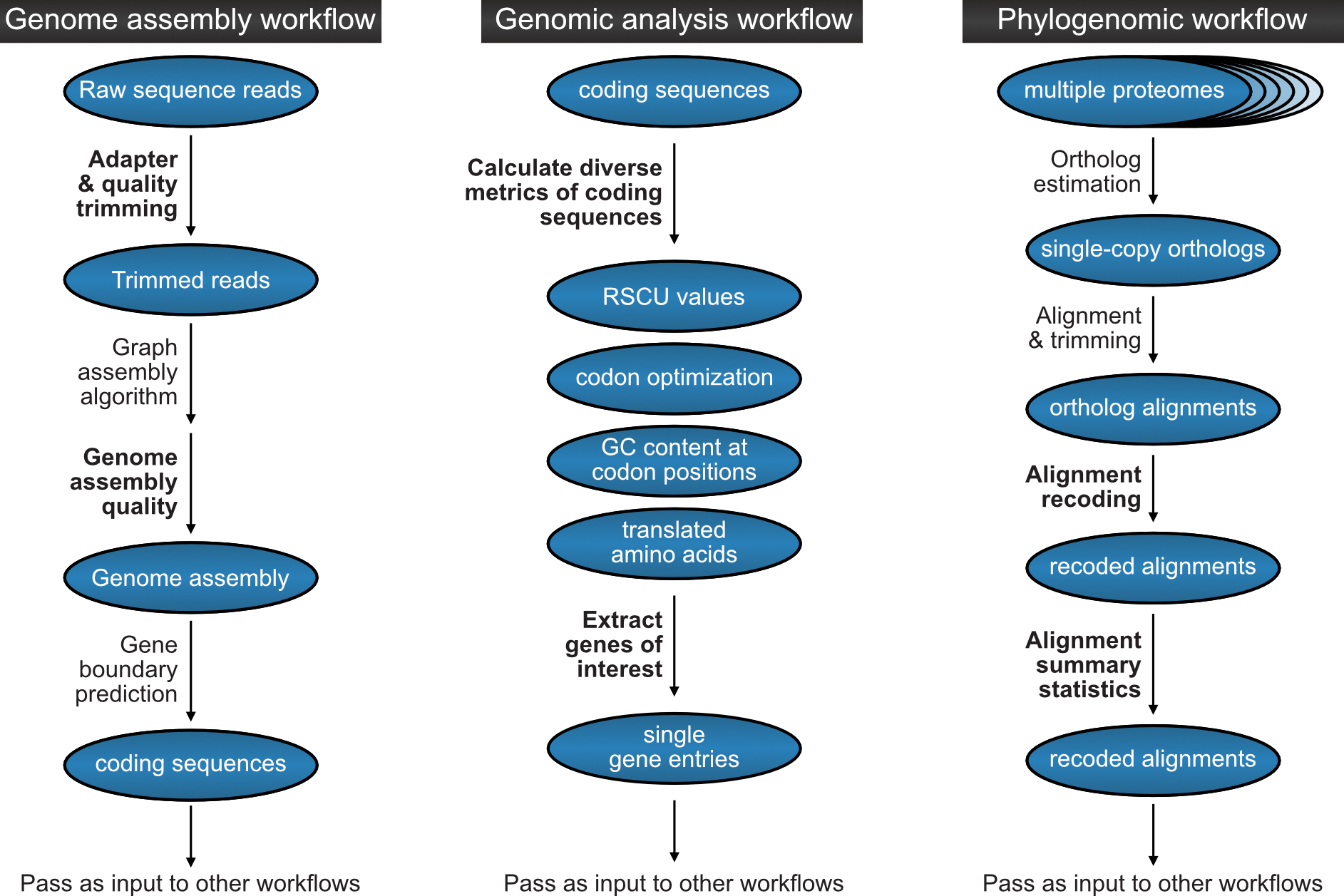
After successful installation, BioKIT can be used in diverse applications. In this figure, we showcase three examples, which cover a small number of total possibilities, where BioKIT can be used. For each example, steps that can be executed by BioKIT are shown in bold.