Evo GoodReads 2022
Gene family evolution and paralog divergence contributes to the evolution of cellular diversity
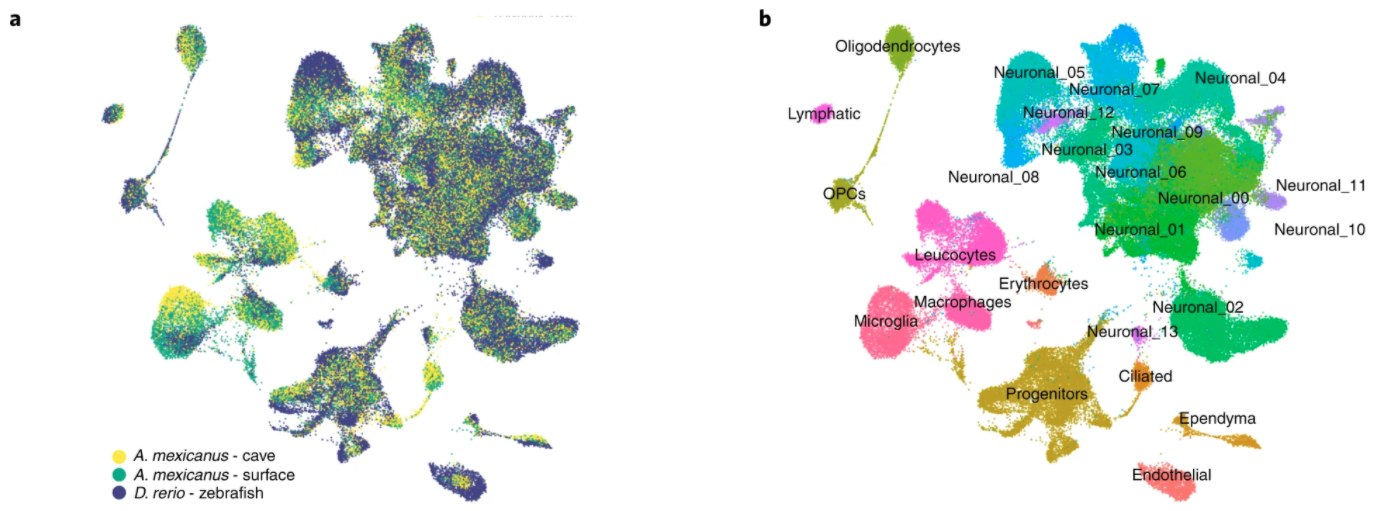 Eukaryotic organisms are made from hundreds of different types of cells, which begs the question: what gave
rise to the spectacular diversity of cells? To begin to shed light on this question, researchers conducted
a focused investigation of brain cell-type diversity by constructing atlases of single-cell expression profiles
for the hypothalamic cells of zebrafish (Danio rerio) and surface and cave morphs of Mexican tetra
(Astyanax mexicanus). These atlases revealed extensive conservation among over 75% of cell-types despite
diverging over 150 million years ago. Notwithstanding these similarities, shared cell-types were observed to differ
in regulation and divergence in paralog expression. Species-specific cells were largely attributed to species-specific
genes and paralog neofunctionalization. Taken together, these findings underscore the impact of
gene family evolution (including the genesis of lineage-specific genes as well as duplication and divergence of
preexisting genes) in the evolution of novel cell-types.
Eukaryotic organisms are made from hundreds of different types of cells, which begs the question: what gave
rise to the spectacular diversity of cells? To begin to shed light on this question, researchers conducted
a focused investigation of brain cell-type diversity by constructing atlases of single-cell expression profiles
for the hypothalamic cells of zebrafish (Danio rerio) and surface and cave morphs of Mexican tetra
(Astyanax mexicanus). These atlases revealed extensive conservation among over 75% of cell-types despite
diverging over 150 million years ago. Notwithstanding these similarities, shared cell-types were observed to differ
in regulation and divergence in paralog expression. Species-specific cells were largely attributed to species-specific
genes and paralog neofunctionalization. Taken together, these findings underscore the impact of
gene family evolution (including the genesis of lineage-specific genes as well as duplication and divergence of
preexisting genes) in the evolution of novel cell-types.
Shafer, M. E. R., Sawh, A. N., and Schier, A. F. (2022).
Gene family evolution underlies cell-type
diversification in the hypothalamus of teleosts. Nat. Ecol. Evol. 6, 63–76. doi:10.1038/s41559-021-01580-3.
Are mutations random? These results suggest they aren't!
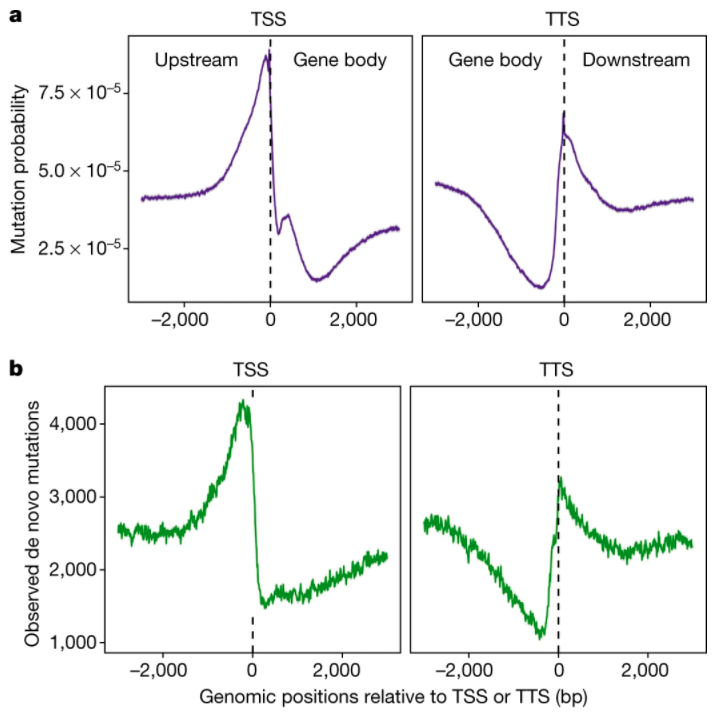 Current wisdom, informed by evolutionary theory, suggests that mutations occur randomly. Here, researchers
test this hypotheis by studying patterns of de novo mutations in Arabidopsis thaliana. In contrast
to expectation, mutations were not observed to occur randomly. Instead, mutations were less frequently found
in protein coding regions. For example, mutations were half as likely to occur in gene bodies and two-thirds
less likely to occur in essential genes. Asymmetric patterns of mutation in the genome can be explained by
epigenomic markers and physical features. This work enabled reseachers to accurately predict mutational
patterns in natural populations of Arabidopsis. Taken together, this work calls into question
the notion that mutation occurs randomly and sets the stage for exciting future research to further our
understanding of mutational biases and evolution.
Current wisdom, informed by evolutionary theory, suggests that mutations occur randomly. Here, researchers
test this hypotheis by studying patterns of de novo mutations in Arabidopsis thaliana. In contrast
to expectation, mutations were not observed to occur randomly. Instead, mutations were less frequently found
in protein coding regions. For example, mutations were half as likely to occur in gene bodies and two-thirds
less likely to occur in essential genes. Asymmetric patterns of mutation in the genome can be explained by
epigenomic markers and physical features. This work enabled reseachers to accurately predict mutational
patterns in natural populations of Arabidopsis. Taken together, this work calls into question
the notion that mutation occurs randomly and sets the stage for exciting future research to further our
understanding of mutational biases and evolution.
Monroe, J. G., Srikant, T., Carbonell-Bejerano, P., Becker, C., Lensink, M., Exposito-Alonso, M., et al. (2022).
Mutation bias reflects natural selection in Arabidopsis thaliana.
Nature. doi:10.1038/s41586-021-04269-6.