About
I'm an evolutionary biologist and bioinformatician investigating how genomes evolve and function, with a particular focus on animals and fungi. My
research explores the evolutionary mechanisms that drive fungal pathogenicity—work that directly impacts our ability to
identify, prevent, and combat fungal infections. By studying the genetic foundations of traits across diverse fungal species, I aim to decode the
fundamental principles governing genome evolution.
Beyond research, I develop computational tools that empower biological discovery. My software
includes ClipKIT and PhyKIT, which streamline phylogenomic
analysis and help researchers conduct more precise evolutionary studies. These tools have been adopted by research groups worldwide, enabling faster and
more accurate scientific discoveries.
Collaboration drives everything I do. The most meaningful scientific breakthroughs happen when diverse minds work together, combining complementary
expertise and perspectives. I'm always excited to explore new partnerships—if you're interested in working together, please
reach out.
Science belongs to everyone. I'm passionate about promoting diversity, equity, and inclusion in STEM through community engagement and accessible science
communication. One way I do this is through Genomely: where genomics and evolution converge—a newsletter
that makes cutting-edge genomics research accessible to everyone. Join thousands of subscribers exploring the fascinating world of genomes and evolution!
Research Interests
Genome Evolution and Function
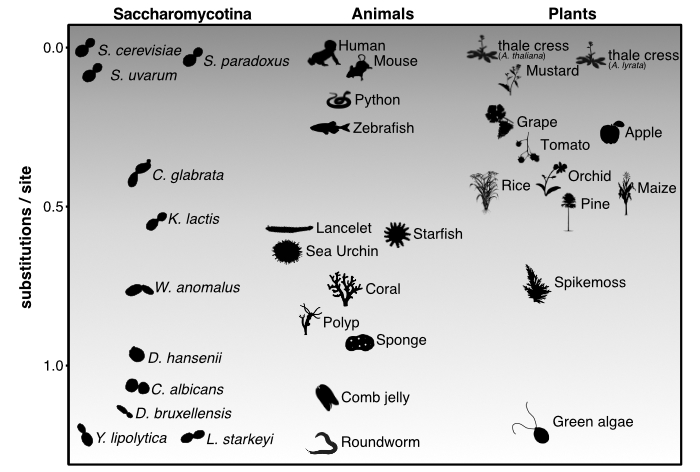 How do genomes evolve and function? This fundamental question drives my research into the Saccharomycotina subphylum of yeasts—one of
nature's most remarkably diverse groups. Budding yeast diversity rivals that of animals and plants combined (see figure from
Shen et al. 2018, Cell), making them ideal for studying evolutionary principles.
Through the Y1000+ initiative, we're sequencing and analyzing over 1,000 budding yeast species,
revealing insights into genome evolution across 400 million years.
How do genomes evolve and function? This fundamental question drives my research into the Saccharomycotina subphylum of yeasts—one of
nature's most remarkably diverse groups. Budding yeast diversity rivals that of animals and plants combined (see figure from
Shen et al. 2018, Cell), making them ideal for studying evolutionary principles.
Through the Y1000+ initiative, we're sequencing and analyzing over 1,000 budding yeast species,
revealing insights into genome evolution across 400 million years.
Our discoveries challenge conventional wisdom. We've shown that genetic networks inferred from evolutionary data capture conserved cellular
architecture and genome function (Steenwyk et al. 2022, Science Advances).
Even more surprising, Hanseniaspora yeasts have lost numerous cell cycle and DNA repair genes once thought essential to all life
(Steenwyk et al. 2019, PLOS Biology). These findings reveal
that genomes are far more dynamic and adaptable than previously imagined.
Steenwyk et al. (2022) Science Advances Steenwyk et al. (2019) PLOS Biology Steenwyk & Rokas (2016) G3
Fungal Pathogenicity
 What transforms a harmless microbe into a deadly pathogen? Filamentous fungi like Aspergillus and Candida exemplify this paradox—
while some species threaten human and plant health, others produce life-saving pharmaceuticals like penicillin. Understanding their evolutionary
histories reveals the molecular switches between harm and help.
What transforms a harmless microbe into a deadly pathogen? Filamentous fungi like Aspergillus and Candida exemplify this paradox—
while some species threaten human and plant health, others produce life-saving pharmaceuticals like penicillin. Understanding their evolutionary
histories reveals the molecular switches between harm and help.
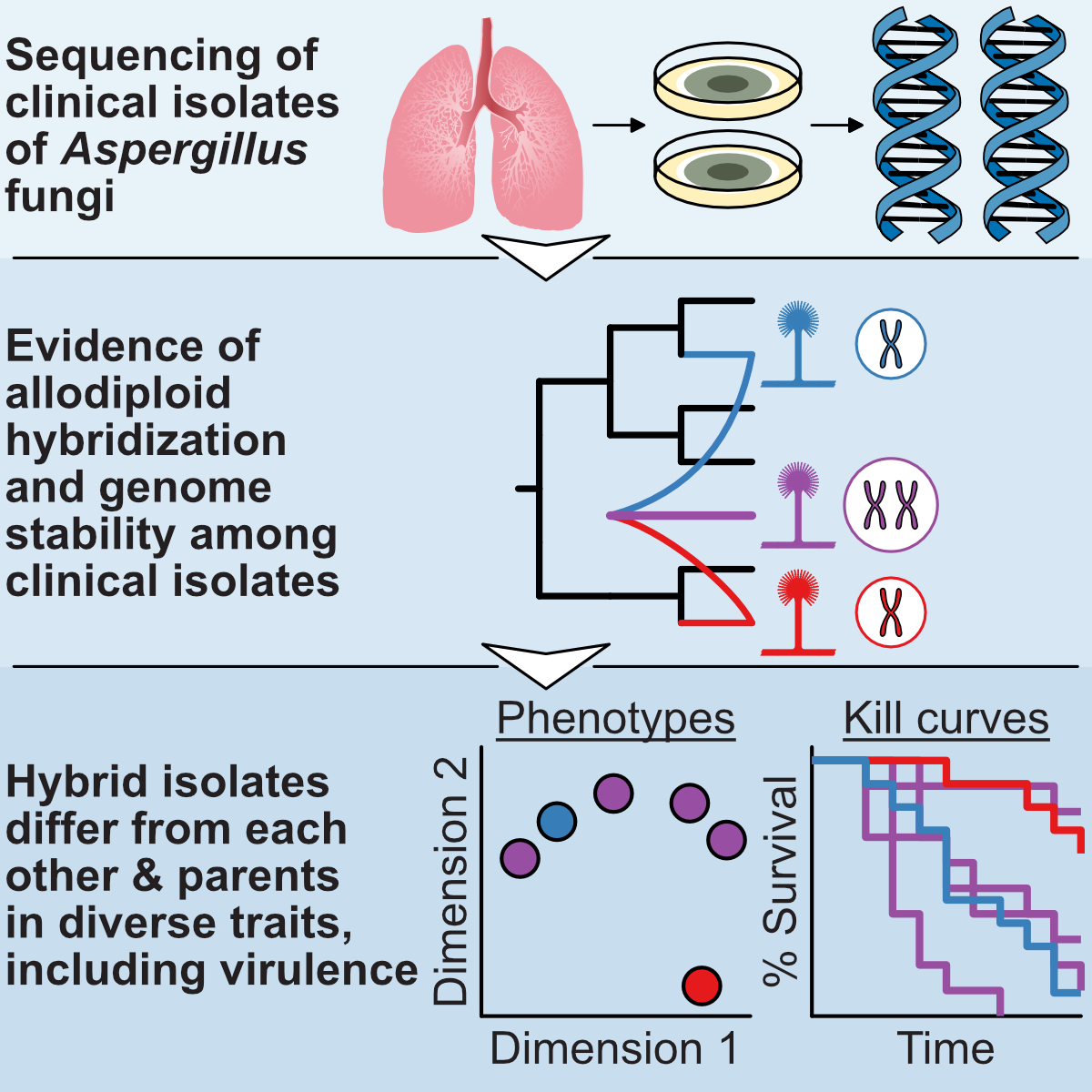
Our research has uncovered unexpected mechanisms driving fungal pathogen evolution. We discovered that Aspergillus latus originated through
allodiploid hybridization, maintaining genome content from both parental species (see figure from
Steenwyk, Lind et al. 2020, Current Biology).
These hybrid pathogens showed unique traits distinct from their parents, suggesting hybridization as a key innovation in pathogen evolution.
Even more intriguingly, we found that supposedly "harmless" Aspergillus species carry virulence genes and display pathogen-like behaviors
(Steenwyk et al. 2020, Genetics). This discovery challenges our fundamental
understanding: what truly makes a pathogen?
Steenwyk et al. (2024) Nature Communications Steenwyk et al. (2023) PLOS Pathogens Steenwyk, Lind et al. (2020) Current Biology
Phylogenomics
 The tree of life tells the story of evolution—but reading that story requires sophisticated methods and careful interpretation. My phylogenomic
research reconstructs evolutionary relationships among species, genes, and biological entities, providing the essential framework for understanding
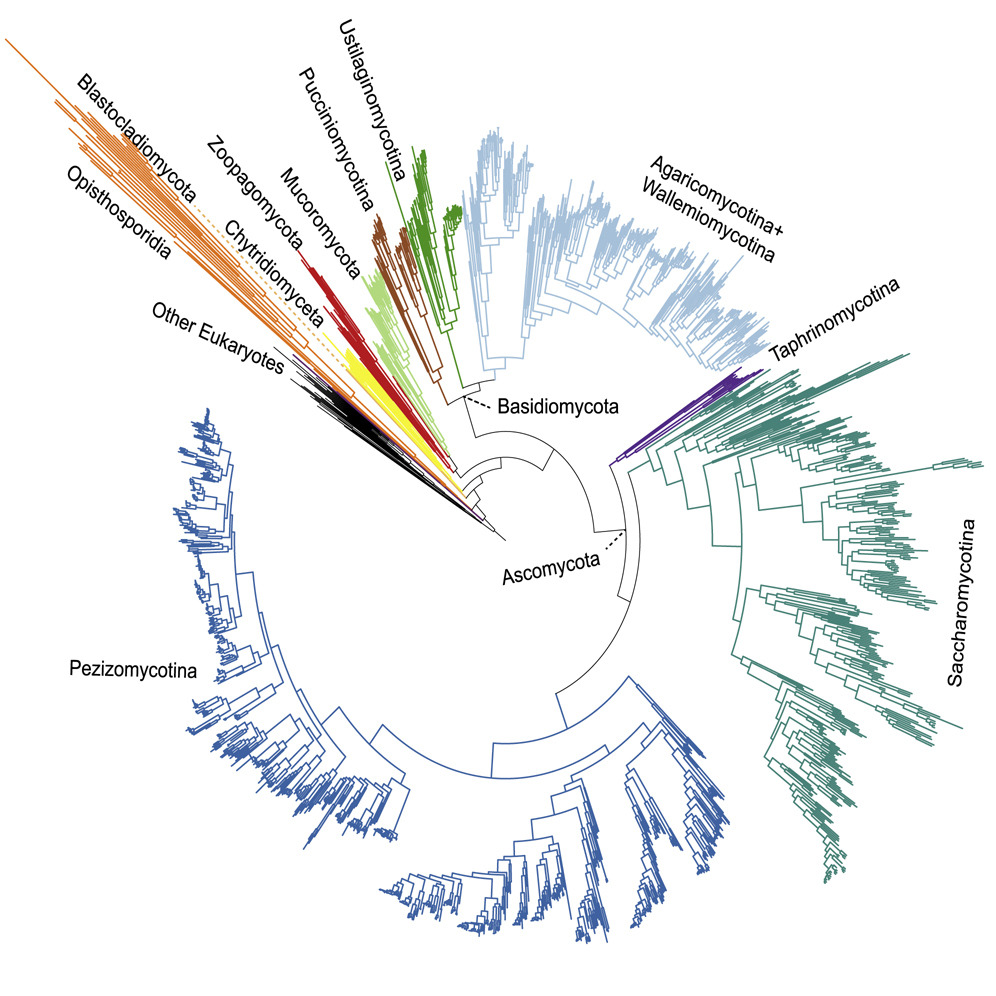
how life diversified. We've mapped fungal evolutionary history (see figure from
Li et al. 2021, Current Biology) and developed novel analytical methods
(Shen et al. 2021, Systematic Biology) that advance the field's capabilities.
The tree of life tells the story of evolution—but reading that story requires sophisticated methods and careful interpretation. My phylogenomic
research reconstructs evolutionary relationships among species, genes, and biological entities, providing the essential framework for understanding
how life diversified. We've mapped fungal evolutionary history (see figure from
Li et al. 2021, Current Biology) and developed novel analytical methods
(Shen et al. 2021, Systematic Biology) that advance the field's capabilities.
One of our most significant contributions addresses a persistent challenge: phylogenetic incongruence, where different datasets yield conflicting
evolutionary hypotheses. Through comprehensive analysis and synthesis, we've characterized this phenomenon and proposed solutions
(Steenwyk et al. 2023, Nature Reviews Genetics).
Our work also places evolutionary events in deep time, revealing when major lineages arose across hundreds of millions of years
(Steenwyk et al. 2019, mBio). These temporal frameworks are crucial
for understanding how evolutionary processes unfold across geological timescales.
Steenwyk et al. (2023) Nature Reviews Genetics Shen et al. (2021) Systematic Biology
Li et al. (2021) Current Biology
Development of Computational Tools
 Modern biology is computational biology. Recognizing that researchers need powerful, accessible tools to unlock biological insights, I develop
software that transforms how scientists analyze evolutionary data. My flagship tool, ClipKIT,
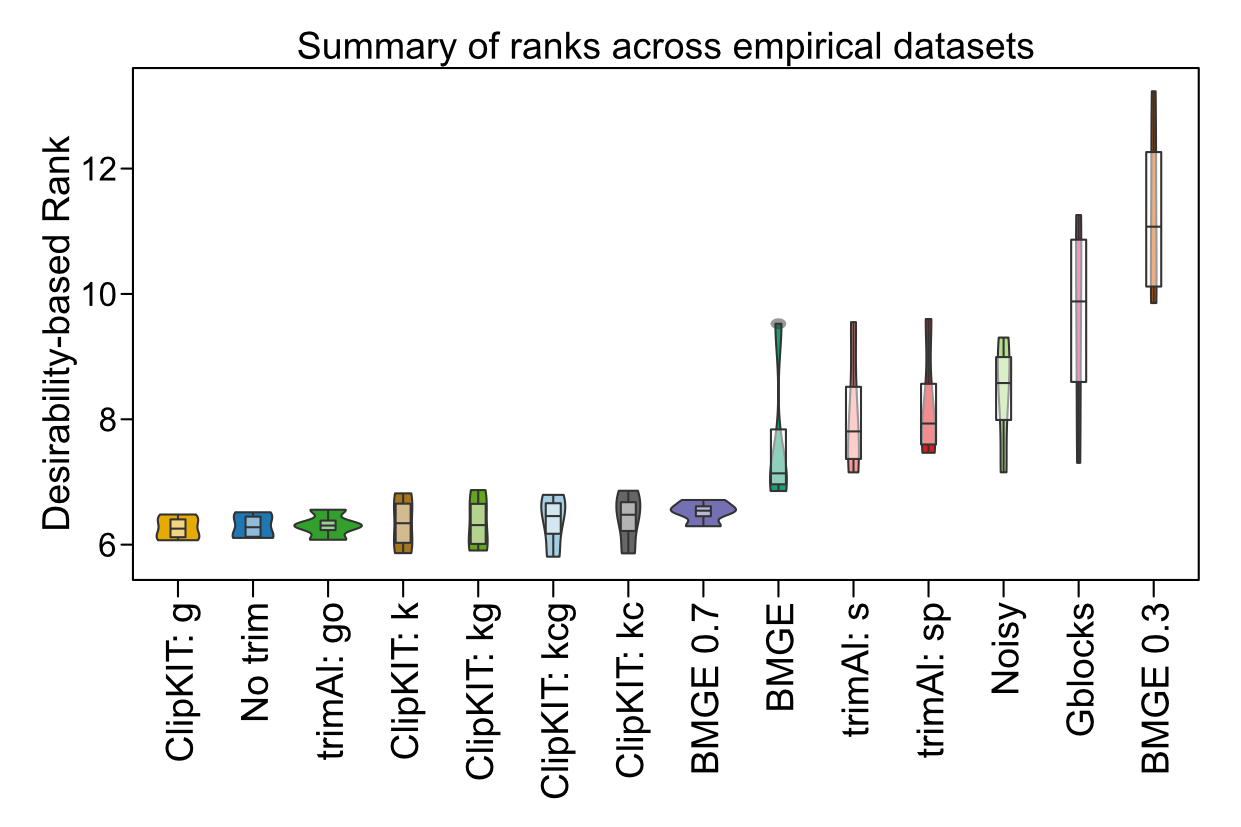
revolutionizes alignment trimming by intelligently retaining phylogenetically informative sites while removing noise. In rigorous testing across
~140,000 alignments, ClipKIT consistently outperformed existing methods (see figure from
Steenwyk et al. 2020, PLOS Biology;
browser application).
Modern biology is computational biology. Recognizing that researchers need powerful, accessible tools to unlock biological insights, I develop
software that transforms how scientists analyze evolutionary data. My flagship tool, ClipKIT,
revolutionizes alignment trimming by intelligently retaining phylogenetically informative sites while removing noise. In rigorous testing across
~140,000 alignments, ClipKIT consistently outperformed existing methods (see figure from
Steenwyk et al. 2020, PLOS Biology;
browser application).
PhyKIT serves as the Swiss Army knife of phylogenomics, enabling researchers to assess data quality,
screen gene function, and detect rapid evolutionary events
(Steenwyk et al. 2021, Bioinformatics). These tools have been adopted
by hundreds of research groups worldwide, accelerating discoveries across evolutionary biology. My complete software portfolio is available on the
software page.
Steenwyk et al. (2022) PLOS Biology Steenwyk et al. (2021) Bioinformatics Steenwyk et al. (2020) PLOS Biology
