Usage
This section covers practical OrthoSNAP usage. For a full worked example, see the tutorial.
OrthoSNAP takes:
a gene tree in Newick format
the FASTA file used to infer that tree
It outputs one FASTA file per inferred SNAP-OG (single-copy orthologous subgroup). Optionally, it can also write a Newick tree per SNAP-OG, an inparalog handling report, and one color-coded subgroup plot for the full input tree.
Basic usage
For most cases, only -f/–fasta and -t/–tree are required:
$ orthosnap -f orthogroup_of_genes.faa -t phylogeny_of_orthogroup_of_genes.tre
Input requirements
FASTA headers and tree tip labels must match.
Taxon and sequence IDs must be separated by the same delimiter in both files.
Default delimiter is | (for example, species_A|gene_001).
Accounting for tree uncertainty
OrthoSNAP can collapse low-support bipartitions before pruning inparalogs.
Default support threshold is 80.
Use -s/–support to change it.
$ orthosnap -f orthogroup_of_genes.faa -t phylogeny_of_orthogroup_of_genes.tre -s 70
Choosing which inparalog to keep
Use -ip/–inparalog_to_keep to select how species-specific inparalogs are resolved.
Supported values:
shortest_seq_len
median_seq_len
longest_seq_len (default)
shortest_branch_len
median_branch_len
longest_branch_len
Examples:
$ orthosnap -f orthogroup_of_genes.faa -t phylogeny_of_orthogroup_of_genes.tre -ip shortest_branch_len
$ orthosnap -f orthogroup_of_genes.faa -t phylogeny_of_orthogroup_of_genes.tre -ip median_seq_len
Inparalog handling report
Use -rih/–report_inparalog_handling to write a tab-delimited report named <input_fasta>.inparalog_report.txt.
Columns are:
SNAP-OG identifier
kept inparalog
trimmed inparalog(s), separated by ;
$ orthosnap -f orthogroup_of_genes.faa -t phylogeny_of_orthogroup_of_genes.tre -rih
Specifying the delimiter
If your headers do not use |, specify the delimiter with -d/–delimiter.
$ orthosnap -f orthogroup_of_genes.faa -t phylogeny_of_orthogroup_of_genes.tre -d -
Plotting SNAP-OG assignments
Use -ps/–plot_snap_ogs to create one figure of the full tree with distinct colors for each inferred SNAP-OG. Default plot format is PNG; choose PDF or SVG with -pf/–plot_format.
$ orthosnap -f orthogroup_of_genes.faa -t phylogeny_of_orthogroup_of_genes.tre -ps
$ orthosnap -f orthogroup_of_genes.faa -t phylogeny_of_orthogroup_of_genes.tre -ps -pf svg
Example output (png):
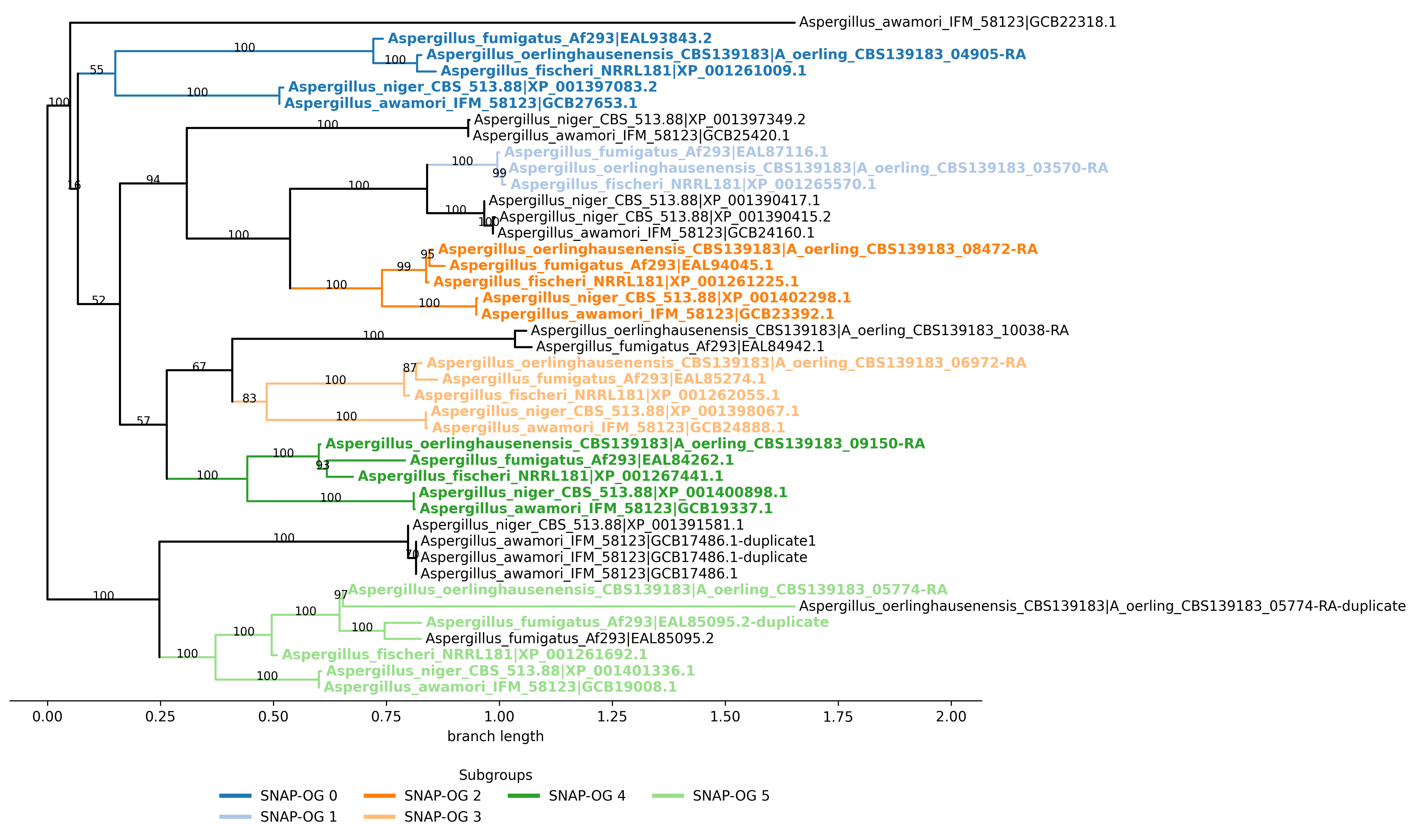
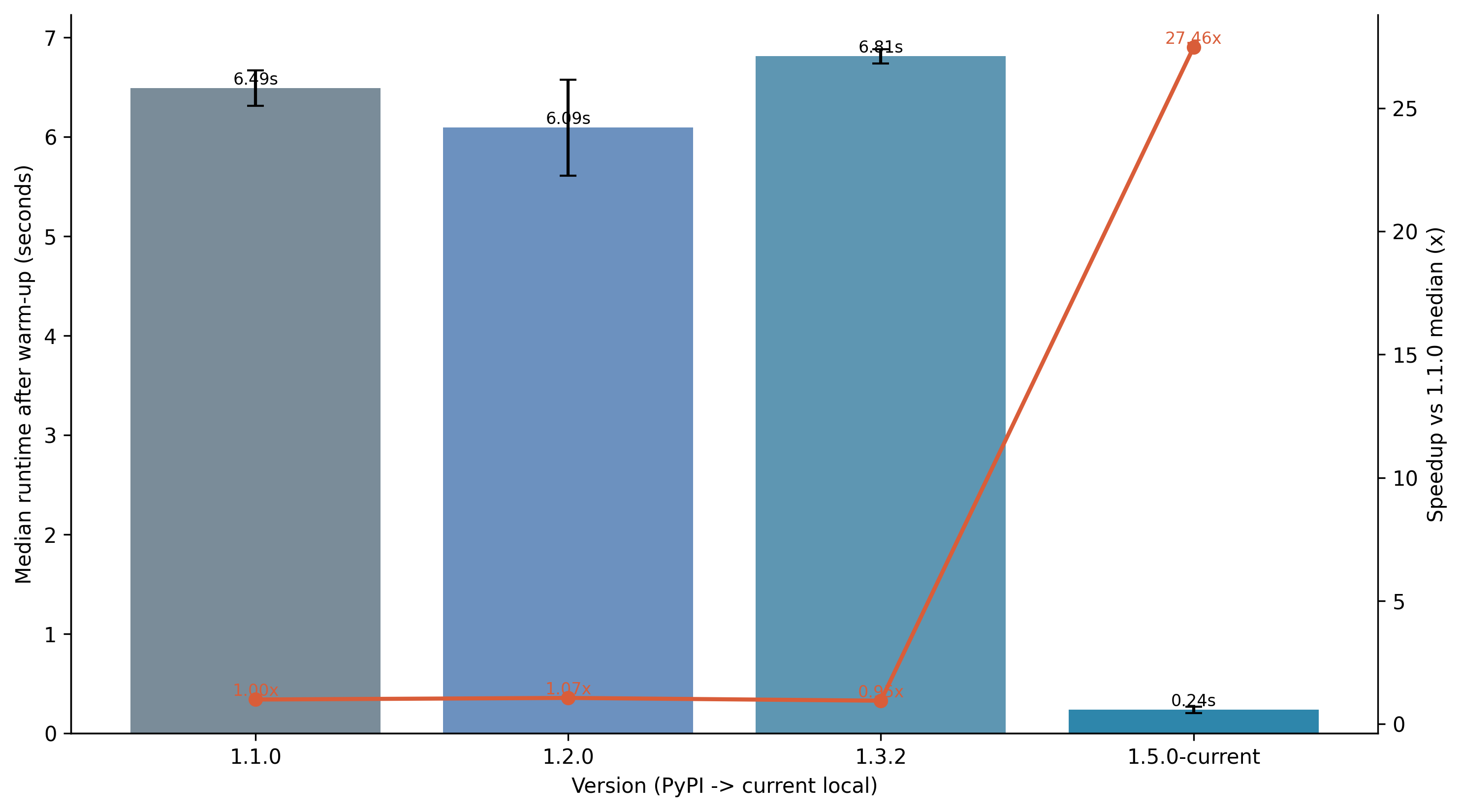
Performance Benchmark
The chart below summarizes benchmarked runtime across selected PyPI releases and the current local version. Runs used a rooted input tree (-r), one warm-up run per version, and three measured runs per version on the same dataset.
Compared versions:
1.1.0 (PyPI baseline)
1.2.0 (PyPI)
1.3.2 (PyPI)
1.5.0-current (local)
All options
Option |
Meaning |
|---|---|
|
Print help message. |
|
Print software version. |
|
Input FASTA file. |
|
Input tree file in Newick format. |
|
Collapse threshold for branch support (default: 80). |
|
Minimum represented taxa for subgroup candidates (default: rounded half of taxa in input FASTA). |
|
Treat input tree as rooted; otherwise midpoint-root it (default: false). |
|
Delimiter between taxon and sequence IDs (default: |
|
Also write SNAP-OG trees in Newick format (default: false). |
|
Rule for keeping one inparalog among species-specific duplicates (default: |
|
Write tab-delimited inparalog handling report (default: false). |
|
Output directory (default: directory containing input FASTA). |
|
Write one color-coded full-tree plot with subgroup labels (default: false). |
|
Output format for subgroup plot ( |
For genome-scale analyses, consider using the same -o/–occupancy value across all gene families to keep SNAP-OG occupancy thresholds consistent.